水稻条纹病毒蛋白与核酸的特性
作者:林含新 林奇田 吴祖建 林奇英 谢联辉
单位:(福建农业大学植物病毒研究所,福建省植物病毒学重点实验室,福州 350002)
关键词:
中国病毒学990409Characterization of Proteins and Nucleic Acid
of Rice Stripe Virus
Lin Hanxin Lin Qitian Wu Zujian Lin Qiying Xie Lianhui
(Institute of Plant Virology of Fujian Agricultural University, Key Laboratory of
, 百拇医药
Plant Virology of Fujian Province, Fuzhou 350002)
Abstract An isolate of rice stripe virus (designated as RSV-YL) was purified. The particles showed to be pleomorphisms under electron microscope, mainly branched filaments of about 80-250 nm in length and about 8 nm in width. There are also some open circular filaments of 3 nm and 8 nm in width, and some filaments of 13 nm in width and 130-190 nm in length. The basic morphism of RSV particles should be filaments of 3 nm in width and various length. By SDS-PAGE analysis, the molecular weight of disease-specific protein (SP) encoded by vRNA4 was 19.9 kDa and that of coat protein (CP) encoded by vcRNA3 was 33.6 kDa. When nucleic acid extracted from the purified RSV was electrophoresed under nondenaturing condition, the size of four dsRNAs (designated as dsRNA1-4 in order of decreasing size) was 4.9×106,2.7×106,2.0×106 and 1.7×106 Da, respectively, and that of four ssRNAs (designated as ssRNA1-4 in order of decreasing size) was 3.0×106,1.2×106,0.9×106 and 0.8×106 Da, respectively. A fifth segment with a size of 0.58×106 Da identified as ssRNA5 associated with the purified virus sometimes. The antiserum against the coat protein further purified by preparative electrophoresis was raised and used to investigate the serological relationships between RSV-CP and RSV-SP, CP and SP of rice grassy stunt virus (RGSV) which is also a member of Tenuivirus. The results showed that RSV-CP had no serological reaction with SP of RSV and PGSV, but could weakly react with antiserum of RGSV-CP, which confirmed that there is distantly evolutionary relationship between RGSV and RSV.
, 百拇医药
Key words Rice stripe virus, Protein, Nucleic acid, Serology
摘 要 提纯的水稻条纹病毒(云南宜良分离物)在电镜下的形态为多型性,但主要是宽8-10 nm,长80-250的分枝丝状体,有些为直径3 nm或8 nm的开环环状体,有些为13 nm宽,130-190 nm长的丝状体,但其基本结构应是直径3 nm、长度不等的丝状体。经聚丙烯酰胺凝胶电泳分析,vRNA4编码的病害特异蛋白(SP)分子量为19.9 kDa,而vcRNA3编码的外壳蛋白(CP)约为33.6 kDa。在非变性条件下,RSV的4条ssRNAs大小分别为3.0×106(ssRNA1)、1.2×106(ssRNA2)、0.9×106(ssRNA3)和0.8×106 Da(ssRNA4),有时出现一条大小为0.58×106 Da的单链RNA(ssRNA5);而4条dsRNAs的分子量分别为4.9×106(dsRNA1)、2.8×106(dsRNA2)、2.0×106(dsRNA3)和1.7×106 Da(dsRNA4)。利用制备电泳分离提纯的外壳蛋白免疫家兔,得到了高特异性的抗血清。A蛋白夹心ELISA检测结果表明,RSV-CP与水稻草状矮化病毒(RGSV)CP抗血清有微弱的反应,但与RSV、RGSV的SP抗血清没有反应,而RSV-CP抗血清与RSV-SP及RGSV的SP、CP都无血清学关系,这个结果表明RGSV与RSV之间在进化上具有一定的亲缘关系。
, http://www.100md.com
关键词 水稻条纹病毒,蛋白,核酸,血清学
Rice stripe virus (RSV) is the typical member of Tenuivirus. The other members are rice grassy stunt virus (RGSV), maize stripe virus (MStV), rice hoja blanca virus (RHBV). RSV causes severe damage to rice production in China, Japan, Korea and former USSR[1].
After several cycles of sucrose density gradient centrifugation, RSV can be separated into three components, M (comprising two subcomponents, M1 and M2), B and NB, which contain circular filaments or filamentous particles with various lengths, four ssRNAs and four dsRNAs were found in association with RSV nucleic acid[2,3]. In infected plant tissues, a large amount of viral -encoded disease-specific protein (SP) accumulate, which are related to the developing of symptoms[4,5]. In Japan, the genome of T isolate has been completely sequenced. RSV genome has an ambisense nature. The SP is encoded by viral-sense RNA4 (vRNA4) and one large open reading frame (ORF) on the viral complementary-sense RNA3 (vcRNA3) encodes coat protein[6-9]. However, the chemical and serological properties of RSV are not clearly characterized in China yet, even though some molecular biological characteristics of RSV has also been studied[10-13]. Here, we report the properties of proteins and nucleic acid of RSV YL isolate in China and the serological relationships among CP, SP of RSV and RGSV.
, 百拇医药
1 Materials and Methods
1.1 Virus and plant materials
RSV isolated from Yiliang County, Yunnan Province, China was maintained in a japonica rice variety (Hexi 28) by transmission via the viruliferous smaller brown planthopper, Laodelphax striaterllus. Infected rice leaves were stored in -70℃ and used for purification.
1.2 Purification of RSV
RSV was purified following the method of Ishikawa et al (1989)[2], except that infected leaf tissues were firstly grounded in liquid nitrogen and that a RPS 65-T rotor (Hitachi) was used for sucrose density gradient centrifugation which was performed for 2.5 h at 101 700 g at 4℃. Virus bands and the zones between the bands were separately collected. Each fraction was diluted with 0.1 mol/L PBS and centrifuged for 1.5 h at 150 000g. The pellet was finally suspended in 0.01 mol/L PBS (containing 0.1% DEPC).
, 百拇医药
1.3 Electron microscopy
Purified RSV was mounted on grids coated with a collodion film for 5 min, then stained with 2% phosphotungastic acid for 5 min, and examined under a JEM-1200 EX electron microscope (JEOL, Japan).
1.4 Preparation and electrophoresis of RSV-RNAs
RNAs were extracted as described by Toriyama (1986)[14], and then analyzed in native 1.5% agrose gels (Bio-Red Chemical). Nucleic acid was also released from the purified RSV by heating for 5-10 min at 55℃ in a solution containing 2% sodium dodecyl sulfate (SDS), 4% glycerol and 1% bromephenol blue just prior to electrophoresis. After electrophoresis, the gels were soaked in distilled water for 10 min and then stained with ethidium bromide.
, http://www.100md.com
ssRNA marker used here was 6 583, 4 981, 2 604, 1 908, 1 383, 955, 623 and 283 bp (Promega Chemical), RDV dsRNAs of 3.29×106, 2.65×106, 2.39×106,2.01×106,1.91×106,1.22×106,1.20×106,0.912×106,0.827×106,0.795×106,0.553×106 and 0.553×106 Da were also used as molecular weight standards.
To determine the properties of RNAs, RNase A (Sigma) treatments in high- and low-salt buffers were carried out according to the method described by Ishikawa et al (1989)[2].
, http://www.100md.com
The photograph of the gels and the calculation of molecular weight were all perfomed on IS-1000 Digital Imaging System (Alpha Innotech Corporation).
1.5 SDS-polyacrylamide gel electrophoresis (PAGE) of proteins
SP was purified following the method of Lin et al (1999)[15]. Purified SP or RSV sample (1 μL) was added with 3 μL loading buffer (1% SDS, 0.5% 2-mer-captoethanol, 2 mmol/L EDTA, 4% glycerol and 0.5% bromephenol blue), then heated at 100 ℃ for 3 min and electrophoresed. After electrophoresis, the gels were stained with coomassie blue and destained, then analyzed on the IS-1000 Digital Imaging System. The markers used were 97.4×103, 66.2×103, 43.0×103, 31.0×103, 20.1×103 and 14.4×103 Da (Shanghai Dong Feng Biochemical Factory).
, http://www.100md.com
1.6 Purification of CP and preparation of antiserum
RSV-CP was separated by preparative electrophoresis. Purified RSV preparation (200 μL, 1.0 mg/mL) was added with 4% glycerol and 0.5% bromephenol blue, then electrophoresed without heating at 100 ℃. After electrophoresis, the gels were soaked in 100 mL 0.2 mol/L KCl. After 5-15min, when the CP band became white, cut out the band and groud it with 1 mL 0.1 mol/L PBS.
A New Zealand white rabbit was immunized with the further purified CP preparation in the leg muscle. A series of four injections with the same amount of antigen, emulsified with an equal volume of Freund′s complete adjuvant were given at weekly intervals. One week after the last intramuscular injection, antigen was injected into the ear vein. Ten days later, the rabbit was sacrificed, and the antiserum was collected and stored at 4 ℃.
, http://www.100md.com
1.7 Protein A sandwich enzyme-linked immunosorbent assay (PAS-ELISA)
ELISA procedure was followed by the method described by Lu et al (1990)[16]. The purified RGSV SP, virus particles and the high specific antiserum against the further purified RGSV SP from gel were kindly provided by our colleague Lin Liming. We are also grateful to Dr. T Omura (National Agricultural Research Center of Japan) for his RGSV antiserum (Philippine isolate).
, 百拇医药
2 Results
2.1 Purification of RSV
After 10%-40% linear sucrose density gradient centrifugation, two distinct visible bands formed. One located in 20% sucrose fraction and the other located in 30% fraction, which was similar to the position of M,B and NB components described by Toriyama (1989)[3]. When analyzed by ultraviolet, we found that a large amount of virus particles also sedimented in 10% and 40% fractions, even in the pellet. When the dissolved pellets from the first ultracentrifugation were centrifuged in second sucrose density gradient, two bands also formed in the same position, and a certain amount of virus was also found to be contained in 10% and 40% fractions. We centrifuged the pooled bands, 10% fraction and 40% fraction, and resuspended the pellets with 50 μL 0.1 mol/L PBS, pH 7.2 (containing 0.1% DEPC), respectively.
, 百拇医药
The purified RSV preparation showed a typical ultraviolet absorption curve, which has a maximum absorbance at 257 nm and a minimum at 239 nm, the OD 260/280 ratio is 1.59 (Fig.1).
Fig.1 Ultraviolet absorption curve of purified RSV preparation
图1 RSV 提纯制剂的紫外吸收曲线
2.2 Electron microscopy
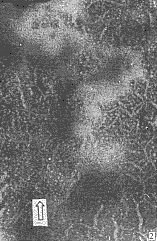
Purified particles showed pleomorphisms under electron microscope. Most particles are branched filaments of about 80-250 nm in length and about 8nm in width. There are also some open circular filaments of 3 nm and 8 nm in width, and some filaments of 13 nm in width and 130-190nm in length (Fig.2).
, 百拇医药
Fig.2 Electron micrograph of RSV preparation stained with 2% phosphotungastic acid
图2 2% 磷钨酸染色的病毒粒体形态
Arrows indicate open circular particles
2.3 Electrophoretic analysis of RSV RNAs
After 1.5% agrose gel electrophoresis, four broad and four sharp bands were observed (Fig.3A). After the gel was treated with RNase A in 2×SSC, the broad bands as well as ssRNA marker disappeared (Fig.3B), indicating that these four bands were ssRNAs. When the gel was treated with RNase A in 0.1×SSC, the sharp bands disappeared as did those of RDV dsRNAs,which meant that the four sharp bands were dsRNAs (Figure not shown). Whenever treated with RNase A in 2×SSC or 0.1×SSC, the DNA bands were always there (Fig.3B).

, 百拇医药
Fig.3 (A) Electrophoresis of RSV RNAs on 1.5% agrose gel
图3 (A) RSV RNAs在1.5%琼脂糖上的电泳图谱
Lane 1:RSV RNAs extracted directly by heating is SDS; Lane 2:ssRNA marker; Lane 3:RDV RNAs;Lane 4,6:RSV RNAs; Lane 5,8:RSV RNAs extracted by SDS and phenol; Lane 7:DNA marker. (B) Digested with RNase A in 2×SSC.
Analyzed by IS-1000 Digital Imaging System, the size of four ssRNAs (designated as ssRNA 1-4 in order of decreasing molecular weight) under nondenaturing condition were approximately 9 147, 3 766, 2 614 and 2 375 bp, respectively, which was similar to the sizes obtained by genome sequencing. Since 1 kb ssRNA is equivalent to 3.3×105 Da, the molecular weights of four ssRNAs were approximately 3.0×106, 1.2×106, 0.9×106 and 0.8×106 Da, respectively. The sizes of four dsRNAs (designated as dsRNA1-4 in order of decreasing molecular weight) were approximately 4.9×106, 2.7×106, 2.0×106 and 1.8×106 Da, respectively.
, 百拇医药
When the pooled bands, 10% and 40% sucrose fractions were ultracentrifuged and resuspended, and then analysed by electrophoresis, we found that these three fractions were all in association with four ssRNAs and four dsRNAs, but the amount of virus is relatively small in 40% fraction (Figure not shown).
The electrophoretic patterns of RNAs extracted by SDS and phenol were compared with those released directly by heating the purified RSV in SDS prior to the electrophoresis. The bands of RNAs extracted by the latter method were much distinctly visible than those of RNAs extracted by the former one (Fig.3A), indicating that RSV RNAs were easy to be degraded and the yield was higher with the latter.
, http://www.100md.com
However, the virus particle we recently purified showed an extra band with a size of 0.58×106 Da on 1.5% agrose gel (Fig.4). When treated with RNase A in 2×SSC, the band disap-peared, which demonstrated that it is ssRNA5 as Ishikawa et al[17] reported.
Fig.4 Electrophoresis of RSV RNAs on 1.5% agrose gel
图4 RSV RNAs在1.5%琼脂糖中的电泳图谱
, 百拇医药
Lane 1-2:RSV RNAs; Lane 3:RDV RNAsFig.5 Electrophoresis of RSV-CP and RSV-SP on polyacrylamide gel
2.4 SDS-PAGE analysis of proteins
RSV CP released from purified virus migrated on polyacrylamide gel as a single band with molecular weight of 33.6 kDa. However, the SP appeared to be a major band and two minor bands on the gel, the size estimated to be 19.9×103, 19.3×103 and 12.1×103 Da, respectively. The two minor bands were believed to be degraded from the major band (Fig.5).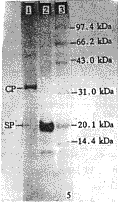
, 百拇医药
图5 外壳蛋白和病害特异性蛋白在聚丙烯酰胺凝胶上的电泳分析
Lane 1:RSV-CP; Lane 2:RSV-SP; Lane 3: Protein marker
2.5 Preparation of antiserum against RSV-CP and investigation of serological relationship
The antiserum against the further purified CP by preparative electrophoresis was highly specific. The titre determined by PAS-ELISA was 6 400 and the optimal working concentration was 500-1 000.
, 百拇医药
PAS-ELISA was used to investigate the serological relationships among RSV-CP and RSV-SP, RGSV-SP, RGSV-CP. The results showed that RSV-CP had no serological reaction with antiserum of RGSV-SP and RSV-SP, but could weakly react with antiserum of RGSV-CP. However, the antiserum of RSV-CP could not react not only with RGSV-SP and RSV-SP, but also with RGSV-CP (Table 1). These results confirmed that RSV has distantly evolutionary relationship with RGSV.
Table 1 Serological relationships amongst RSV-CP, SP and RGSV-CP, SP as shown by absorbent values (A405) in PAS-ELISA
, 百拇医药
表1 PAS-ELISA 测定RSV-CP与RSV-SP、RGSV-CP、SP之间的血清学关系 Antisera
Antigen
G-SP
(H)
G-CP
(H)
S-SP
(H)
S-CP
(H)
G-Sap
(H)
, 百拇医药
S-Sap
(H)
CK
S-CP
0.80
(0.50)
0.58
(0.43)
0.84
(0.55)
1.61*
(0.50)
0.82
, 百拇医药
(0.55)
1.97*
(0.30)
0.31
G-CP
0.66
(0.48)
2.30*
(0.57)
0.92
(0.54)
1.16*
, 百拇医药
(0.54)
2.30*
(0.60)
0.69
(0.50)
0.32
G-SP
1.71*
(0.36)
0.40
(0.39)
0.38
, 百拇医药
(0.37)
0.39
(0.36)
2.03*
(0.36)
0.39
(0.36)
0.35
Note: G-SP:RGSV-SP;G-CP:purified RGSV particles;S-SP:RSV-SP;S-CP:purified RSV particles;G-Sap:RGSV leaf sap;S-Sap:RSV leaf sap;H:healthy leaf sap;* :positive reaction;CK:blank control.
, 百拇医药
3 Discussion
RSV particles show pleomorphism under electron microscope.In different components, different researchers have reported branched filaments,circular filaments,filaments or rod-like particles of 3 nm or 8 nm in width and various length[2,3,17-22]. But the basic structure of RSV particles must be filaments of 3 nm in width and various length, which can form into superhelix confirmation of 8 nm in width[4]. In this investigation, RSV particles also show various morphisms as branched filaments, circular filaments and filaments of various width (Fig.2). We believe that RSV particles must be filaments of 3 nm or 8 nm in width and the branched filaments and filaments of over 8 nm in width must be formed by partly or completely overlay of virus particles. The circular filaments may be due to the complementarity between 5′- and 3′-terminal sequences of each segment under certain ion environment[23]. The pleomorphism of RSV particles is due to the ion concentrations and types of buffer used in purification[24].
, 百拇医药
After two cycles of sucrose density gradient centrifugation, RSV particles still distributed in each fraction, all containing four dsRNAs and four ssRNAs (Figure not shown). These results suggested that RSV particles become entangled easily and are difficult to separate completely into single components. This entanglement was believed to be the filamentous nature of the virus, and may be affected by virus concentration, purity and some other factors[3].
, http://www.100md.com The sizes of RSV dsRNAs and ssRNAs in this investigation were similar to those of RSV T and M strains already described by Toriyama et al (1989)[2] and Ishikawa et al (1989)[3]. We found that an extra ssRNA5 with a size of 0.58×106 Da was associated with purified virus particles sometimes. In Japan, this fifth segment was not found in RSV particles of T isolate. To the M isolate, Ishikawa et al also did not report in their first paper about characteristics of nucleic acid in 1989[3], but they soon identified this segment in their second paper in 1989[17]. Our results also suggested that this fifth segment is not isolate- or strain-specific, since all 7 RSV isolates including 3 strains of China contain ssRNA5 (unpublished data). Recently, ssRNA5 was shown to be a subgenomic RNA with the part of the vRNA4 sequence, but lacking the 3′ half sequence[25]. Now, the questions that why this subgenomic RNA appears irregularly, when and how much it produces in the life cycle of virus are deserved of further researching.
, 百拇医药
There have been many reports on serological relationships among members of Tenuivirus[22,26-31], which could be summarized in table 2 combined with our results. There were also some reports on serological relationship between RSV and RGSV. But those reports were not full-scaled and sometimes conflicted each other. For example, Chen et al[32] reported that there was no serological reaction between RSV and RGSV. But in another paper, they found that antiserum of RSV could react with RGSV[33]. Hibino et al[26] believed that there was distantly serological relationship between RSV and RGSV. In this investigation, we also found that there was weakly reaction between RSV and antiserum of RGSV, but no reaction between antiserum of RSV and RGSV. Recently, Toriyama compared the nucleotide sequence identity among Tenuivirus. The results showed that the RSV CP gene shares only 20.9% amino acid identity with RGSV and 65.2%, 48.4% identities with MStV, RHBV, respectively, and MStV CP gene has 48.4% identity with RHBV[34,35]. But among these four members, there are only serological relationships between RSV-CP and RGSV-CP, MStV-CP (see table 2). So the serological reaction between two proteins not only depends on sequence identity at amino acid level, but may be related to secondary conformation of proteins.
, 百拇医药
Table 2 The serological relationships among members of Tenuivirus
表2 纤细病毒组各成员之间的血清学关系 Antisera
Antigen
MStV
-CP
MStV
-SP
RSV-
CP
RSV-
SP
, 百拇医药
RGSV-
CP
RGSV-
SP
RHBV-
CP
RHBV-
SP
EHBV-
CP
EHBV-
SP
MStV-CP
, http://www.100md.com
-
?
-
?
-
?
-
?
MStV-SP
-
?
, 百拇医药
-
?
?
?
-
-
-
RSV-CP
-
-
+
, 百拇医药
-
-
?
?
?
RSV-SP
-
-
-
-
+
?
?
, 百拇医药
?
?
RGSV-CP
-
?
+
-
-
-
?
?
?
RGSV-SP
, http://www.100md.com
?
?
-
+
-
?
?
?
?
RHBV-CP
-
?
-
, 百拇医药
?
-
?
-
?
RHBV-SP
-
-
?
?
?
, http://www.100md.com
?
-
?
*: reference from Gingery et al (1983); Falk et al (1987); Hibino et al (1985); Morales et al (1983, 1985); Horita et al (1983) and Miranda et al (1995). EHBV: echinochloa hoja blanca virus, a tentative member of Tenuivirus.
+ represents intensity of serological reaction. Reaction intensity increases with the number of +; represents no serological relationship; represents indefinite serological relationship.
, 百拇医药
Acknowledgements This work was supported by the National Natural Science Foundation (item number 39670489) and Natural Science Foundation of Fujian Province (item number C97031).
*国家自然科学基金和福建省自然科学基金资助课题
References
1 Murphy FA, Fanaquet CM, Biship DHL et al. Classification and nomenclature of viruses: Sixth Report of International Committee on Taxonomy of Viruses. Arch Virol, 1995, (Suppl 10):316
, 百拇医药
2 Toriyama S, Watanabe Y. Characterization of single- and double-stranded RNAs in particles of rice stripe virus. J Gen Virol, 1989,70:505-511
3 Ishikawa K, Omura T, Tsuchzaki T. Association of double- and single-stranded RNAs with each four components of rice stripe virus. Ann Phytopathol Soc Jpn, 1989,35(3):315-323
4 Toriyama S. Rice stripe virus: prototype of a new group of viruses that replicated in plants and insects. Microbiol Sci, 1986, 3:347-351
, http://www.100md.com
5 Lin Qitian, Lin Hanxin, Wu Zujian et al. Accumulations of coat protein and disease-specific protein of rice stripe virus in its host. Journal of Fujian Agricultural University, 1998,27(3): 322-326
6 Zhu Y, Hayakawa T, Toriyama S. Complete nucleotide sequence of RNA3 of rice stripe virus: an ambisense coding strategy. J Gen Virol, 1991,72:763-767
7 Zhu Y, Hayakawa T, Toriyama S. Complete nucleotide sequence of RNA4 of rice stripe virus isolate T and comparison with another isolate and with maize stripe virus. J Gen Virol, 1992, 73:1309-1312
, http://www.100md.com
8 Takahashi M, Toriyama S, Hamamatsu C et al. Nucleotide sequence and possible ambisence coding strategy of rice stripe virus RNA segment 2. J Gen Virol, 1993,74:769-773
9 Toriyama S, Takahashi M, Sano Y et al. Nucleotide sequence of RNA1: the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J Gen Virol, 1994,75:3569-3579
10 Hong Yiguo, Pei Meiyun, Wang Xiaofeng et al. Study on molecular biology of rice stripe virus Ⅰ. Molecular weight, amino acid composition and N termini of coat protein. Chinese Science Bulletin, 1990,35(16): 1265-1267 (in Chinese)
, http://www.100md.com
11 Qui Bingsheng, Wang Jinfang, Tian Po. Study on molecular biology of rice stripe virus Ⅱ. Synthesis, cloning and expression of coat protein gene. Chinese Science Bulletin, 1991,36(20): 1578-1581 (in Chinese)
12 Wang Jinfang, Qui Bingsheng, Tian Po. Study on molecular biology of rice stripe virus Ⅲ. Sequence analysis of coat protein gene. Virologica Sinica, 1992,7(4): 463-466 (in Chinese)
13 Wang Xiaohong, Yie Yin, Wang Suyan et al. Clone, sequence analysis and expression in prokaryote of vRNA2 ORF of rice stripe virus genome. Chinese Science Bulletin, 1996,42(4): 438-441 (in Chinese)
, http://www.100md.com
14 Toriyama S. An RNA-dependent RNA polymerase associated with the filamentous nucleoproteins of rice stripe virus. J Gen Virol, 1986, 67:1247-1255
15 Lin Hanxin, Lin Qitian, Wu Zujian et al. Purification, serology and characterization of disease-specific protein of rice stripe virus. Virologica Sinica, 1999,14(3):222-229
16 Lu Jiayu, Zhang Chengliang, Zhang Zuofang et al. Studies on the detection of plant viruses by using protein A-ELISA. Plant Quarantine, 1990,4(3):161-163 (in Chinese)
, 百拇医药
17 Iskikawa K, Omura T, Hibino H. Morphological characteristics of rice stripe virus. J Gen Virol, 1989, 70:505-511
18 Koganezawa H, Doi Y, Yora K. Purification of rice stripe virus. Ann Phytopathol Soc Jpn, 1975,41:148-154
19 Koganezawa H. Purification and properties of rice stripe virus. In: Symposium on Virus Diseases of Tropical Crops. Tropical Agricultural Research Series, 1977. (10):151-154
, 百拇医药
20 Toriyama S. Three ribonucleic acids associated with rice stripe virus. Ann Phytopathol Soc Jpn, 1982, 48:482-489
21 Toriyama S. Characterization of rice stripe virus: a heavy component carrying infectivity. J Gen Virol, 1982, 61:187-195
22 Horita M, Tsushima S, Ugeda I et al. Serology of rice stripe virus. Mem Fac Agric Hokaido Univ, 1983,13:551
23 Takahashi M, Toriyama S, Hamamatsu C et al. Complementarity between the 5′- and 3′-terminal sequences of rice stripe virus RNAs. J Gen Virol, 1990,71:1817-1882
, http://www.100md.com
24 Toriyama S, Suzuki Y, Goto Y et al. Effects of salts on RNA polymerase activity and conformation of filamentous nucleoproteins of rice stripe virus. Ann Phytopathol Soc Jpn, 1994,60:555-562
25 Ishikawa K, Omura T. Characteristics of segment 5 of rice stripe virus which encodes the non-capsid protein (NCP) (abstract). Ann Phytopathol Soc Jpn, 1996,62:340
26 Hibino H, Usugi T, Omura T et al. Rice grassy stunt virus: a planthopper-borne circular filaments. Phytopathology, 1985,75:894-899
, http://www.100md.com
27 Gingery RE, Nault LR, Yamashita S. Relationship between maize stripe and rice stripe virus. J Gen Virol, 1983,64:1765-1770
28 Miranda GJ, Koganezawa H. Identification, purification and serology detection of major noncapsid protein of rice grassy stunt virus. Phytopathology, 1995, 85:1530-1533
29 Falk BW, Tsai JH. Serological and biochemical properties of capsid and major noncapsid proteins of maize stripe, rice hoja blanca, and echinochloa hoja blanca viruses. Phytopathology, 1987,77(2):196-201
, 百拇医药
30 Morales FJ, Niessen AI. Association of spiral filamentous virus-like particles with rice hoja blanca. Phytopathlogy, 1983,73:971
31 Morales FJ, Niessen AI. Rice hoja blanca virus, In: Description of Plant Viruses, No.199, Association of Applied biologists, National Vegetable Research Station, Wellesbourne, Warwich, U.K. 1985.
32 Chen CC, Huang WK. Purification, partial characterization and serological properties of rice stripe virus. Bulletin of Taichung District Agricultural Improvement Station, 1993,41:33-42
, http://www.100md.com
33 Chen CC, Huang WL, Ko WH et al. Purification, characterization and serological analysis of rice wilted stunt virus.In: Chiu Renjong et al eds. Council of Agricultural Plant Protection Series No.1. Proceedings of the Symposium on Plant Virus and Virus-like Diseases. Taiwang. 1993. 177-191
34 Toriyama S, Kimishima T, Takahashi M. The proteins encoded by rice grassy stunt virus RNA5 and RNA6 are distantly related to the corresponding proteins of other members of the genus Tenuivirus. J Gen Virol, 1997,78:2355-2363
35 Toriyama S, Kimishima T, Takahashi M et al. The complete nucleotide sequence of the rice grassy stunt virus genome and genomic comparisons with viruses of the genus Tenuivirus. J Gen Virol, 1998, 79:2051-2058
*收稿日期:1998-08-11,修回日期:1999-02-04, 百拇医药
单位:(福建农业大学植物病毒研究所,福建省植物病毒学重点实验室,福州 350002)
关键词:
中国病毒学990409Characterization of Proteins and Nucleic Acid
of Rice Stripe Virus
Lin Hanxin Lin Qitian Wu Zujian Lin Qiying Xie Lianhui
(Institute of Plant Virology of Fujian Agricultural University, Key Laboratory of
, 百拇医药
Plant Virology of Fujian Province, Fuzhou 350002)
Abstract An isolate of rice stripe virus (designated as RSV-YL) was purified. The particles showed to be pleomorphisms under electron microscope, mainly branched filaments of about 80-250 nm in length and about 8 nm in width. There are also some open circular filaments of 3 nm and 8 nm in width, and some filaments of 13 nm in width and 130-190 nm in length. The basic morphism of RSV particles should be filaments of 3 nm in width and various length. By SDS-PAGE analysis, the molecular weight of disease-specific protein (SP) encoded by vRNA4 was 19.9 kDa and that of coat protein (CP) encoded by vcRNA3 was 33.6 kDa. When nucleic acid extracted from the purified RSV was electrophoresed under nondenaturing condition, the size of four dsRNAs (designated as dsRNA1-4 in order of decreasing size) was 4.9×106,2.7×106,2.0×106 and 1.7×106 Da, respectively, and that of four ssRNAs (designated as ssRNA1-4 in order of decreasing size) was 3.0×106,1.2×106,0.9×106 and 0.8×106 Da, respectively. A fifth segment with a size of 0.58×106 Da identified as ssRNA5 associated with the purified virus sometimes. The antiserum against the coat protein further purified by preparative electrophoresis was raised and used to investigate the serological relationships between RSV-CP and RSV-SP, CP and SP of rice grassy stunt virus (RGSV) which is also a member of Tenuivirus. The results showed that RSV-CP had no serological reaction with SP of RSV and PGSV, but could weakly react with antiserum of RGSV-CP, which confirmed that there is distantly evolutionary relationship between RGSV and RSV.
, 百拇医药
Key words Rice stripe virus, Protein, Nucleic acid, Serology
摘 要 提纯的水稻条纹病毒(云南宜良分离物)在电镜下的形态为多型性,但主要是宽8-10 nm,长80-250的分枝丝状体,有些为直径3 nm或8 nm的开环环状体,有些为13 nm宽,130-190 nm长的丝状体,但其基本结构应是直径3 nm、长度不等的丝状体。经聚丙烯酰胺凝胶电泳分析,vRNA4编码的病害特异蛋白(SP)分子量为19.9 kDa,而vcRNA3编码的外壳蛋白(CP)约为33.6 kDa。在非变性条件下,RSV的4条ssRNAs大小分别为3.0×106(ssRNA1)、1.2×106(ssRNA2)、0.9×106(ssRNA3)和0.8×106 Da(ssRNA4),有时出现一条大小为0.58×106 Da的单链RNA(ssRNA5);而4条dsRNAs的分子量分别为4.9×106(dsRNA1)、2.8×106(dsRNA2)、2.0×106(dsRNA3)和1.7×106 Da(dsRNA4)。利用制备电泳分离提纯的外壳蛋白免疫家兔,得到了高特异性的抗血清。A蛋白夹心ELISA检测结果表明,RSV-CP与水稻草状矮化病毒(RGSV)CP抗血清有微弱的反应,但与RSV、RGSV的SP抗血清没有反应,而RSV-CP抗血清与RSV-SP及RGSV的SP、CP都无血清学关系,这个结果表明RGSV与RSV之间在进化上具有一定的亲缘关系。
, http://www.100md.com
关键词 水稻条纹病毒,蛋白,核酸,血清学
Rice stripe virus (RSV) is the typical member of Tenuivirus. The other members are rice grassy stunt virus (RGSV), maize stripe virus (MStV), rice hoja blanca virus (RHBV). RSV causes severe damage to rice production in China, Japan, Korea and former USSR[1].
After several cycles of sucrose density gradient centrifugation, RSV can be separated into three components, M (comprising two subcomponents, M1 and M2), B and NB, which contain circular filaments or filamentous particles with various lengths, four ssRNAs and four dsRNAs were found in association with RSV nucleic acid[2,3]. In infected plant tissues, a large amount of viral -encoded disease-specific protein (SP) accumulate, which are related to the developing of symptoms[4,5]. In Japan, the genome of T isolate has been completely sequenced. RSV genome has an ambisense nature. The SP is encoded by viral-sense RNA4 (vRNA4) and one large open reading frame (ORF) on the viral complementary-sense RNA3 (vcRNA3) encodes coat protein[6-9]. However, the chemical and serological properties of RSV are not clearly characterized in China yet, even though some molecular biological characteristics of RSV has also been studied[10-13]. Here, we report the properties of proteins and nucleic acid of RSV YL isolate in China and the serological relationships among CP, SP of RSV and RGSV.
, 百拇医药
1 Materials and Methods
1.1 Virus and plant materials
RSV isolated from Yiliang County, Yunnan Province, China was maintained in a japonica rice variety (Hexi 28) by transmission via the viruliferous smaller brown planthopper, Laodelphax striaterllus. Infected rice leaves were stored in -70℃ and used for purification.
1.2 Purification of RSV
RSV was purified following the method of Ishikawa et al (1989)[2], except that infected leaf tissues were firstly grounded in liquid nitrogen and that a RPS 65-T rotor (Hitachi) was used for sucrose density gradient centrifugation which was performed for 2.5 h at 101 700 g at 4℃. Virus bands and the zones between the bands were separately collected. Each fraction was diluted with 0.1 mol/L PBS and centrifuged for 1.5 h at 150 000g. The pellet was finally suspended in 0.01 mol/L PBS (containing 0.1% DEPC).
, 百拇医药
1.3 Electron microscopy
Purified RSV was mounted on grids coated with a collodion film for 5 min, then stained with 2% phosphotungastic acid for 5 min, and examined under a JEM-1200 EX electron microscope (JEOL, Japan).
1.4 Preparation and electrophoresis of RSV-RNAs
RNAs were extracted as described by Toriyama (1986)[14], and then analyzed in native 1.5% agrose gels (Bio-Red Chemical). Nucleic acid was also released from the purified RSV by heating for 5-10 min at 55℃ in a solution containing 2% sodium dodecyl sulfate (SDS), 4% glycerol and 1% bromephenol blue just prior to electrophoresis. After electrophoresis, the gels were soaked in distilled water for 10 min and then stained with ethidium bromide.
, http://www.100md.com
ssRNA marker used here was 6 583, 4 981, 2 604, 1 908, 1 383, 955, 623 and 283 bp (Promega Chemical), RDV dsRNAs of 3.29×106, 2.65×106, 2.39×106,2.01×106,1.91×106,1.22×106,1.20×106,0.912×106,0.827×106,0.795×106,0.553×106 and 0.553×106 Da were also used as molecular weight standards.
To determine the properties of RNAs, RNase A (Sigma) treatments in high- and low-salt buffers were carried out according to the method described by Ishikawa et al (1989)[2].
, http://www.100md.com
The photograph of the gels and the calculation of molecular weight were all perfomed on IS-1000 Digital Imaging System (Alpha Innotech Corporation).
1.5 SDS-polyacrylamide gel electrophoresis (PAGE) of proteins
SP was purified following the method of Lin et al (1999)[15]. Purified SP or RSV sample (1 μL) was added with 3 μL loading buffer (1% SDS, 0.5% 2-mer-captoethanol, 2 mmol/L EDTA, 4% glycerol and 0.5% bromephenol blue), then heated at 100 ℃ for 3 min and electrophoresed. After electrophoresis, the gels were stained with coomassie blue and destained, then analyzed on the IS-1000 Digital Imaging System. The markers used were 97.4×103, 66.2×103, 43.0×103, 31.0×103, 20.1×103 and 14.4×103 Da (Shanghai Dong Feng Biochemical Factory).
, http://www.100md.com
1.6 Purification of CP and preparation of antiserum
RSV-CP was separated by preparative electrophoresis. Purified RSV preparation (200 μL, 1.0 mg/mL) was added with 4% glycerol and 0.5% bromephenol blue, then electrophoresed without heating at 100 ℃. After electrophoresis, the gels were soaked in 100 mL 0.2 mol/L KCl. After 5-15min, when the CP band became white, cut out the band and groud it with 1 mL 0.1 mol/L PBS.
A New Zealand white rabbit was immunized with the further purified CP preparation in the leg muscle. A series of four injections with the same amount of antigen, emulsified with an equal volume of Freund′s complete adjuvant were given at weekly intervals. One week after the last intramuscular injection, antigen was injected into the ear vein. Ten days later, the rabbit was sacrificed, and the antiserum was collected and stored at 4 ℃.
, http://www.100md.com
1.7 Protein A sandwich enzyme-linked immunosorbent assay (PAS-ELISA)
ELISA procedure was followed by the method described by Lu et al (1990)[16]. The purified RGSV SP, virus particles and the high specific antiserum against the further purified RGSV SP from gel were kindly provided by our colleague Lin Liming. We are also grateful to Dr. T Omura (National Agricultural Research Center of Japan) for his RGSV antiserum (Philippine isolate).
, 百拇医药
2 Results
2.1 Purification of RSV
After 10%-40% linear sucrose density gradient centrifugation, two distinct visible bands formed. One located in 20% sucrose fraction and the other located in 30% fraction, which was similar to the position of M,B and NB components described by Toriyama (1989)[3]. When analyzed by ultraviolet, we found that a large amount of virus particles also sedimented in 10% and 40% fractions, even in the pellet. When the dissolved pellets from the first ultracentrifugation were centrifuged in second sucrose density gradient, two bands also formed in the same position, and a certain amount of virus was also found to be contained in 10% and 40% fractions. We centrifuged the pooled bands, 10% fraction and 40% fraction, and resuspended the pellets with 50 μL 0.1 mol/L PBS, pH 7.2 (containing 0.1% DEPC), respectively.
, 百拇医药
The purified RSV preparation showed a typical ultraviolet absorption curve, which has a maximum absorbance at 257 nm and a minimum at 239 nm, the OD 260/280 ratio is 1.59 (Fig.1).

Fig.1 Ultraviolet absorption curve of purified RSV preparation
图1 RSV 提纯制剂的紫外吸收曲线
2.2 Electron microscopy
Purified particles showed pleomorphisms under electron microscope. Most particles are branched filaments of about 80-250 nm in length and about 8nm in width. There are also some open circular filaments of 3 nm and 8 nm in width, and some filaments of 13 nm in width and 130-190nm in length (Fig.2).


, 百拇医药
Fig.2 Electron micrograph of RSV preparation stained with 2% phosphotungastic acid
图2 2% 磷钨酸染色的病毒粒体形态
Arrows indicate open circular particles
2.3 Electrophoretic analysis of RSV RNAs
After 1.5% agrose gel electrophoresis, four broad and four sharp bands were observed (Fig.3A). After the gel was treated with RNase A in 2×SSC, the broad bands as well as ssRNA marker disappeared (Fig.3B), indicating that these four bands were ssRNAs. When the gel was treated with RNase A in 0.1×SSC, the sharp bands disappeared as did those of RDV dsRNAs,which meant that the four sharp bands were dsRNAs (Figure not shown). Whenever treated with RNase A in 2×SSC or 0.1×SSC, the DNA bands were always there (Fig.3B).


, 百拇医药
Fig.3 (A) Electrophoresis of RSV RNAs on 1.5% agrose gel
图3 (A) RSV RNAs在1.5%琼脂糖上的电泳图谱
Lane 1:RSV RNAs extracted directly by heating is SDS; Lane 2:ssRNA marker; Lane 3:RDV RNAs;Lane 4,6:RSV RNAs; Lane 5,8:RSV RNAs extracted by SDS and phenol; Lane 7:DNA marker. (B) Digested with RNase A in 2×SSC.
Analyzed by IS-1000 Digital Imaging System, the size of four ssRNAs (designated as ssRNA 1-4 in order of decreasing molecular weight) under nondenaturing condition were approximately 9 147, 3 766, 2 614 and 2 375 bp, respectively, which was similar to the sizes obtained by genome sequencing. Since 1 kb ssRNA is equivalent to 3.3×105 Da, the molecular weights of four ssRNAs were approximately 3.0×106, 1.2×106, 0.9×106 and 0.8×106 Da, respectively. The sizes of four dsRNAs (designated as dsRNA1-4 in order of decreasing molecular weight) were approximately 4.9×106, 2.7×106, 2.0×106 and 1.8×106 Da, respectively.
, 百拇医药
When the pooled bands, 10% and 40% sucrose fractions were ultracentrifuged and resuspended, and then analysed by electrophoresis, we found that these three fractions were all in association with four ssRNAs and four dsRNAs, but the amount of virus is relatively small in 40% fraction (Figure not shown).
The electrophoretic patterns of RNAs extracted by SDS and phenol were compared with those released directly by heating the purified RSV in SDS prior to the electrophoresis. The bands of RNAs extracted by the latter method were much distinctly visible than those of RNAs extracted by the former one (Fig.3A), indicating that RSV RNAs were easy to be degraded and the yield was higher with the latter.
, http://www.100md.com
However, the virus particle we recently purified showed an extra band with a size of 0.58×106 Da on 1.5% agrose gel (Fig.4). When treated with RNase A in 2×SSC, the band disap-peared, which demonstrated that it is ssRNA5 as Ishikawa et al[17] reported.

Fig.4 Electrophoresis of RSV RNAs on 1.5% agrose gel
图4 RSV RNAs在1.5%琼脂糖中的电泳图谱
, 百拇医药
Lane 1-2:RSV RNAs; Lane 3:RDV RNAsFig.5 Electrophoresis of RSV-CP and RSV-SP on polyacrylamide gel
2.4 SDS-PAGE analysis of proteins
RSV CP released from purified virus migrated on polyacrylamide gel as a single band with molecular weight of 33.6 kDa. However, the SP appeared to be a major band and two minor bands on the gel, the size estimated to be 19.9×103, 19.3×103 and 12.1×103 Da, respectively. The two minor bands were believed to be degraded from the major band (Fig.5).

, 百拇医药
图5 外壳蛋白和病害特异性蛋白在聚丙烯酰胺凝胶上的电泳分析
Lane 1:RSV-CP; Lane 2:RSV-SP; Lane 3: Protein marker
2.5 Preparation of antiserum against RSV-CP and investigation of serological relationship
The antiserum against the further purified CP by preparative electrophoresis was highly specific. The titre determined by PAS-ELISA was 6 400 and the optimal working concentration was 500-1 000.
, 百拇医药
PAS-ELISA was used to investigate the serological relationships among RSV-CP and RSV-SP, RGSV-SP, RGSV-CP. The results showed that RSV-CP had no serological reaction with antiserum of RGSV-SP and RSV-SP, but could weakly react with antiserum of RGSV-CP. However, the antiserum of RSV-CP could not react not only with RGSV-SP and RSV-SP, but also with RGSV-CP (Table 1). These results confirmed that RSV has distantly evolutionary relationship with RGSV.
Table 1 Serological relationships amongst RSV-CP, SP and RGSV-CP, SP as shown by absorbent values (A405) in PAS-ELISA
, 百拇医药
表1 PAS-ELISA 测定RSV-CP与RSV-SP、RGSV-CP、SP之间的血清学关系 Antisera
Antigen
G-SP
(H)
G-CP
(H)
S-SP
(H)
S-CP
(H)
G-Sap
(H)
, 百拇医药
S-Sap
(H)
CK
S-CP
0.80
(0.50)
0.58
(0.43)
0.84
(0.55)
1.61*
(0.50)
0.82
, 百拇医药
(0.55)
1.97*
(0.30)
0.31
G-CP
0.66
(0.48)
2.30*
(0.57)
0.92
(0.54)
1.16*
, 百拇医药
(0.54)
2.30*
(0.60)
0.69
(0.50)
0.32
G-SP
1.71*
(0.36)
0.40
(0.39)
0.38
, 百拇医药
(0.37)
0.39
(0.36)
2.03*
(0.36)
0.39
(0.36)
0.35
Note: G-SP:RGSV-SP;G-CP:purified RGSV particles;S-SP:RSV-SP;S-CP:purified RSV particles;G-Sap:RGSV leaf sap;S-Sap:RSV leaf sap;H:healthy leaf sap;* :positive reaction;CK:blank control.
, 百拇医药
3 Discussion
RSV particles show pleomorphism under electron microscope.In different components, different researchers have reported branched filaments,circular filaments,filaments or rod-like particles of 3 nm or 8 nm in width and various length[2,3,17-22]. But the basic structure of RSV particles must be filaments of 3 nm in width and various length, which can form into superhelix confirmation of 8 nm in width[4]. In this investigation, RSV particles also show various morphisms as branched filaments, circular filaments and filaments of various width (Fig.2). We believe that RSV particles must be filaments of 3 nm or 8 nm in width and the branched filaments and filaments of over 8 nm in width must be formed by partly or completely overlay of virus particles. The circular filaments may be due to the complementarity between 5′- and 3′-terminal sequences of each segment under certain ion environment[23]. The pleomorphism of RSV particles is due to the ion concentrations and types of buffer used in purification[24].
, 百拇医药
After two cycles of sucrose density gradient centrifugation, RSV particles still distributed in each fraction, all containing four dsRNAs and four ssRNAs (Figure not shown). These results suggested that RSV particles become entangled easily and are difficult to separate completely into single components. This entanglement was believed to be the filamentous nature of the virus, and may be affected by virus concentration, purity and some other factors[3].
, http://www.100md.com The sizes of RSV dsRNAs and ssRNAs in this investigation were similar to those of RSV T and M strains already described by Toriyama et al (1989)[2] and Ishikawa et al (1989)[3]. We found that an extra ssRNA5 with a size of 0.58×106 Da was associated with purified virus particles sometimes. In Japan, this fifth segment was not found in RSV particles of T isolate. To the M isolate, Ishikawa et al also did not report in their first paper about characteristics of nucleic acid in 1989[3], but they soon identified this segment in their second paper in 1989[17]. Our results also suggested that this fifth segment is not isolate- or strain-specific, since all 7 RSV isolates including 3 strains of China contain ssRNA5 (unpublished data). Recently, ssRNA5 was shown to be a subgenomic RNA with the part of the vRNA4 sequence, but lacking the 3′ half sequence[25]. Now, the questions that why this subgenomic RNA appears irregularly, when and how much it produces in the life cycle of virus are deserved of further researching.
, 百拇医药
There have been many reports on serological relationships among members of Tenuivirus[22,26-31], which could be summarized in table 2 combined with our results. There were also some reports on serological relationship between RSV and RGSV. But those reports were not full-scaled and sometimes conflicted each other. For example, Chen et al[32] reported that there was no serological reaction between RSV and RGSV. But in another paper, they found that antiserum of RSV could react with RGSV[33]. Hibino et al[26] believed that there was distantly serological relationship between RSV and RGSV. In this investigation, we also found that there was weakly reaction between RSV and antiserum of RGSV, but no reaction between antiserum of RSV and RGSV. Recently, Toriyama compared the nucleotide sequence identity among Tenuivirus. The results showed that the RSV CP gene shares only 20.9% amino acid identity with RGSV and 65.2%, 48.4% identities with MStV, RHBV, respectively, and MStV CP gene has 48.4% identity with RHBV[34,35]. But among these four members, there are only serological relationships between RSV-CP and RGSV-CP, MStV-CP (see table 2). So the serological reaction between two proteins not only depends on sequence identity at amino acid level, but may be related to secondary conformation of proteins.
, 百拇医药
Table 2 The serological relationships among members of Tenuivirus
表2 纤细病毒组各成员之间的血清学关系 Antisera
Antigen
MStV
-CP
MStV
-SP
RSV-
CP
RSV-
SP
, 百拇医药
RGSV-
CP
RGSV-
SP
RHBV-
CP
RHBV-
SP
EHBV-
CP
EHBV-
SP
MStV-CP

, http://www.100md.com
-

?
-
?
-
?
-
?
MStV-SP
-

?
, 百拇医药
-
?
?
?
-
-
-
RSV-CP

-

-
+
, 百拇医药
-
-
?
?
?
RSV-SP
-
-
-

-
+
?
?
, 百拇医药
?
?
RGSV-CP
-
?
+
-

-
-
?
?
?
RGSV-SP
, http://www.100md.com
?
?
-
+
-

?
?
?
?
RHBV-CP
-
?
-
, 百拇医药
?
-
?

-

?
RHBV-SP
-
-
?
?
?
, http://www.100md.com
?
-

?

*: reference from Gingery et al (1983); Falk et al (1987); Hibino et al (1985); Morales et al (1983, 1985); Horita et al (1983) and Miranda et al (1995). EHBV: echinochloa hoja blanca virus, a tentative member of Tenuivirus.
+ represents intensity of serological reaction. Reaction intensity increases with the number of +; represents no serological relationship; represents indefinite serological relationship.
, 百拇医药
Acknowledgements This work was supported by the National Natural Science Foundation (item number 39670489) and Natural Science Foundation of Fujian Province (item number C97031).
*国家自然科学基金和福建省自然科学基金资助课题
References
1 Murphy FA, Fanaquet CM, Biship DHL et al. Classification and nomenclature of viruses: Sixth Report of International Committee on Taxonomy of Viruses. Arch Virol, 1995, (Suppl 10):316
, 百拇医药
2 Toriyama S, Watanabe Y. Characterization of single- and double-stranded RNAs in particles of rice stripe virus. J Gen Virol, 1989,70:505-511
3 Ishikawa K, Omura T, Tsuchzaki T. Association of double- and single-stranded RNAs with each four components of rice stripe virus. Ann Phytopathol Soc Jpn, 1989,35(3):315-323
4 Toriyama S. Rice stripe virus: prototype of a new group of viruses that replicated in plants and insects. Microbiol Sci, 1986, 3:347-351
, http://www.100md.com
5 Lin Qitian, Lin Hanxin, Wu Zujian et al. Accumulations of coat protein and disease-specific protein of rice stripe virus in its host. Journal of Fujian Agricultural University, 1998,27(3): 322-326
6 Zhu Y, Hayakawa T, Toriyama S. Complete nucleotide sequence of RNA3 of rice stripe virus: an ambisense coding strategy. J Gen Virol, 1991,72:763-767
7 Zhu Y, Hayakawa T, Toriyama S. Complete nucleotide sequence of RNA4 of rice stripe virus isolate T and comparison with another isolate and with maize stripe virus. J Gen Virol, 1992, 73:1309-1312
, http://www.100md.com
8 Takahashi M, Toriyama S, Hamamatsu C et al. Nucleotide sequence and possible ambisence coding strategy of rice stripe virus RNA segment 2. J Gen Virol, 1993,74:769-773
9 Toriyama S, Takahashi M, Sano Y et al. Nucleotide sequence of RNA1: the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J Gen Virol, 1994,75:3569-3579
10 Hong Yiguo, Pei Meiyun, Wang Xiaofeng et al. Study on molecular biology of rice stripe virus Ⅰ. Molecular weight, amino acid composition and N termini of coat protein. Chinese Science Bulletin, 1990,35(16): 1265-1267 (in Chinese)
, http://www.100md.com
11 Qui Bingsheng, Wang Jinfang, Tian Po. Study on molecular biology of rice stripe virus Ⅱ. Synthesis, cloning and expression of coat protein gene. Chinese Science Bulletin, 1991,36(20): 1578-1581 (in Chinese)
12 Wang Jinfang, Qui Bingsheng, Tian Po. Study on molecular biology of rice stripe virus Ⅲ. Sequence analysis of coat protein gene. Virologica Sinica, 1992,7(4): 463-466 (in Chinese)
13 Wang Xiaohong, Yie Yin, Wang Suyan et al. Clone, sequence analysis and expression in prokaryote of vRNA2 ORF of rice stripe virus genome. Chinese Science Bulletin, 1996,42(4): 438-441 (in Chinese)
, http://www.100md.com
14 Toriyama S. An RNA-dependent RNA polymerase associated with the filamentous nucleoproteins of rice stripe virus. J Gen Virol, 1986, 67:1247-1255
15 Lin Hanxin, Lin Qitian, Wu Zujian et al. Purification, serology and characterization of disease-specific protein of rice stripe virus. Virologica Sinica, 1999,14(3):222-229
16 Lu Jiayu, Zhang Chengliang, Zhang Zuofang et al. Studies on the detection of plant viruses by using protein A-ELISA. Plant Quarantine, 1990,4(3):161-163 (in Chinese)
, 百拇医药
17 Iskikawa K, Omura T, Hibino H. Morphological characteristics of rice stripe virus. J Gen Virol, 1989, 70:505-511
18 Koganezawa H, Doi Y, Yora K. Purification of rice stripe virus. Ann Phytopathol Soc Jpn, 1975,41:148-154
19 Koganezawa H. Purification and properties of rice stripe virus. In: Symposium on Virus Diseases of Tropical Crops. Tropical Agricultural Research Series, 1977. (10):151-154
, 百拇医药
20 Toriyama S. Three ribonucleic acids associated with rice stripe virus. Ann Phytopathol Soc Jpn, 1982, 48:482-489
21 Toriyama S. Characterization of rice stripe virus: a heavy component carrying infectivity. J Gen Virol, 1982, 61:187-195
22 Horita M, Tsushima S, Ugeda I et al. Serology of rice stripe virus. Mem Fac Agric Hokaido Univ, 1983,13:551
23 Takahashi M, Toriyama S, Hamamatsu C et al. Complementarity between the 5′- and 3′-terminal sequences of rice stripe virus RNAs. J Gen Virol, 1990,71:1817-1882
, http://www.100md.com
24 Toriyama S, Suzuki Y, Goto Y et al. Effects of salts on RNA polymerase activity and conformation of filamentous nucleoproteins of rice stripe virus. Ann Phytopathol Soc Jpn, 1994,60:555-562
25 Ishikawa K, Omura T. Characteristics of segment 5 of rice stripe virus which encodes the non-capsid protein (NCP) (abstract). Ann Phytopathol Soc Jpn, 1996,62:340
26 Hibino H, Usugi T, Omura T et al. Rice grassy stunt virus: a planthopper-borne circular filaments. Phytopathology, 1985,75:894-899
, http://www.100md.com
27 Gingery RE, Nault LR, Yamashita S. Relationship between maize stripe and rice stripe virus. J Gen Virol, 1983,64:1765-1770
28 Miranda GJ, Koganezawa H. Identification, purification and serology detection of major noncapsid protein of rice grassy stunt virus. Phytopathology, 1995, 85:1530-1533
29 Falk BW, Tsai JH. Serological and biochemical properties of capsid and major noncapsid proteins of maize stripe, rice hoja blanca, and echinochloa hoja blanca viruses. Phytopathology, 1987,77(2):196-201
, 百拇医药
30 Morales FJ, Niessen AI. Association of spiral filamentous virus-like particles with rice hoja blanca. Phytopathlogy, 1983,73:971
31 Morales FJ, Niessen AI. Rice hoja blanca virus, In: Description of Plant Viruses, No.199, Association of Applied biologists, National Vegetable Research Station, Wellesbourne, Warwich, U.K. 1985.
32 Chen CC, Huang WK. Purification, partial characterization and serological properties of rice stripe virus. Bulletin of Taichung District Agricultural Improvement Station, 1993,41:33-42
, http://www.100md.com
33 Chen CC, Huang WL, Ko WH et al. Purification, characterization and serological analysis of rice wilted stunt virus.In: Chiu Renjong et al eds. Council of Agricultural Plant Protection Series No.1. Proceedings of the Symposium on Plant Virus and Virus-like Diseases. Taiwang. 1993. 177-191
34 Toriyama S, Kimishima T, Takahashi M. The proteins encoded by rice grassy stunt virus RNA5 and RNA6 are distantly related to the corresponding proteins of other members of the genus Tenuivirus. J Gen Virol, 1997,78:2355-2363
35 Toriyama S, Kimishima T, Takahashi M et al. The complete nucleotide sequence of the rice grassy stunt virus genome and genomic comparisons with viruses of the genus Tenuivirus. J Gen Virol, 1998, 79:2051-2058
*收稿日期:1998-08-11,修回日期:1999-02-04, 百拇医药